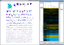
clusterMaker2 is the Cytoscape 3 version of the clusterMaker plugin. clusterMaker2 provides several clustering algorithms for clustering data within columns as well as clustering nodes within a network. This version also provides support for two new algorithms: Fuzzy C-Means and a new "Fuzzifier". In addition to providing clustering algorithms, clusterMaker2 provides heatmap visualization of both node data and edge data as well as the ability to create new networks based on the results of a clustering algorithm.
Current node attribute algorithms:
* AutoSOME
* Create correlation network
* Hierarchical
* K-Means
* K-Medoid
* HOPACH
* PAM
The current network partitioning algorithms are:
* Affinity propagation
* AutoSOME
* Fast Greedy (run remotely)
* Cluster "Fuzzifier"
* Connected Components
* Community Clustering (GLay)
* Fuzzy C-Means Clustering
* Infomap (run remotely)
* Leiden (run remotely)
* Label propagation (run remotely)
* Leading Eigenvector (run remotely)
* MCL
* MCODE
* Multilevel cluster (run remotely)
* SCPS (Spectral Clustering of Protein Sequences)
* Transitivity Clustering
four post-cluster filters:
* Best Neighbor filter
* Cutting Edge filter
* Density filter
* Haircut filter
five cluster ranking algorithms:
* Additive sum
* Multiply sum
* Page Rank
* Page Rank with priors
* Hyperlink induced topic search
and 9 dimensionality reduction techniques:
* Isomap (run remotely)
* Local Linear Embedding (run remotely)
* MDS (Multi-Dimensional Scaling - run remotely)
* PCA
* PCoA
* Spectral (run remotely)
* t-SNE
* t-SNE (run remotely)
* UMAP (run remotely)
In addition, clusterMaker2 provides 5 ways to visualize the data:
* Create New Network from Attribute
* Create new Network from clusters
* JTree TreeView
* JTree KnnView
* JTree HeatMap view (unclustered)