## *Core App*: aMatReader
### Note: This app is pre-installed in Cytoscape 3.9 or later
**_You should not download this app_ from the store yourself. If Cytoscape detects that a new version is available in the App Store, it will notify you as it's starting and will give you a chance to download it directly.**
**_You should not download this app_ if your Cytoscape is earlier than 3.9. An earlier version of this app is pre-installed in your Cytoscape, and this app cannot replace it.**
### Import Adjacency Matrices into Cytoscape
aMatReader is a Cytoscape 3 app that augments the Cytoscape network readers by adding the ability to read in adjacency matrix file(s). If importing with drag'n'drop or the File>Import>Network, the input file must have the ".mat" or ".adj" extension to avoid conflict with other readers. Missing data is assumed to indicate that no edge exists between two nodes. There is no assumption that the nodes defined in the header row and the nodes defined in each data row are in the same order or are matching. Try importing multiple matrix files simultaneously!
Here are a few example matrix formats:
commented, tab delimited, empty values
#Simple tab-delimited adjacency matrix
Node1 Node2 Node3 Node4
Node1 1 4
Node2 1 3 3
Node3 4 3 1
Node4 3 1
OR column prefix, comma separated, NA for null values, row order changed
Rows,sim.Node1,sim.Node2,sim.Node3,sim.Node4
Node2,1,NA,3,3
Node4,NA,3,1,NA
Node1,NA,1,4,NA
Node3,4,3,NA,1
OR space delimited, no column names, zero values
Node1 0 1 4 0
Node2 1 0 3 3
Node3 4 3 0 1
Node4 0 3 1 0
OR no row names, bar delimited
Node1|Node2|Node3|Node4
0|1|4|0
1|0|3|3
4|3|0|1
0|3|1|0
All of the above matrices will result in the same network.
Important things to note:
- Lines starting with `#` are considered comments and ignored
- The value at 0x0 is ignored if row and column names are present
- Supported delimiters: tab, bar, space, and comma
- Only integer and floating point values are considered edge weights
- Column name prefixes can be removed (common in R)
- Row names and column names can be in any order if both names are provided
- otherwise:
- matrix must be square
- row/column names are assumed to be in the same order
*More sample matrix formats can be found in the samples folder, in the parent directory of the [http://github.com/BrettJSettle/aMatReader Github repository]
**Note: aMatReader does NOT handle flat matrix format `source target value` as it is already handled by Cytoscape standard import**
### How to use aMatReader
There are 3 ways to import adjacency matrices via Cytoscape, as well as two CyREST endpoints and a Pypi package
1. The File Menu (1 at a time)
- File > Import > Network > File
- Select a single file in the dialog
- Set parameters for matrix import
2. Drag'n'Drop
- Drag file(s) into the Cytoscape Network Panel
- Dialog will update to import files one at a time
3. Apps Menu
- Apps > AMatReader > Import Matrix file(s)
- Select any files in the dialog
- Dialog will update to import files one at a time
The parameter dialog will peek at each file and attempt to predict some of the import parameters for you. The dialog should look like this:
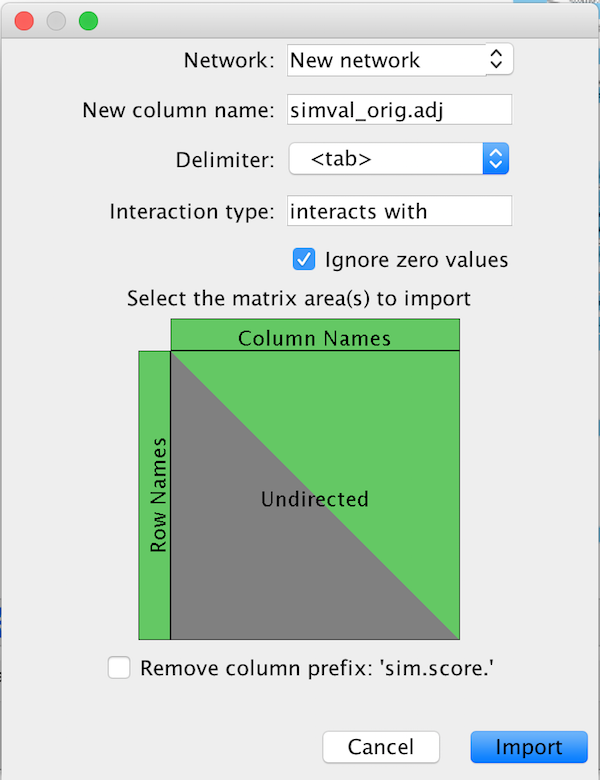
The dialog handles one file at a time, and allows the user to import adjacency matrices as a new network, or as new edge attributes for an existing network.
**There are two CyREST endpoints for aMatReader**
1. `/v1/aMatReader/import` : import matrix files to a new network
2. `/v1/aMatReader/extend/{SUID}` : add edge attributes to an existing network
Both endpoints take similar parameters to those available in the dialog. However, if you are importing files that expect different parameters (eg delimiters), two separate CyREST calls must be made.
**PyCyAMatReader**
Try using the Pypi package to import matrices and files in your scripts. Better documentation on the package is available at [https://github.com/BrettJSettle/aMatReader]
For more help, issues, or ideas regarding aMatReader, contact Brett Settle at the Ideker lab (bsettle@ucsd.edu)
